
-

����ͨ��
����ץס�����Ƽ�
����������
DNA������ʼ��H2A.Z��С���ѡ����ʶ�����
�����壺 �� �� С �� ʱ�䣺2023��08��03�� ��Դ���п�Ժ
�༭�Ƽ���
���������Ŵ���Ϣ�ľ�ȷ���ݶ������ķ��ܺͽ���������Ҫ���ߵ��������DNA������Ҫȷ���״�DNA�������Ŵ���Ϣȷ���ݸ��Ӵ���������ʼλ�����ȷѡ�������е���Ҫ���ڡ������о��������ڶ�ϸ�������к��鵰�ױ���H2A.Z�ĺ�С��ͨ������鵰���������ת��øSUV420H1�ٽ��鵰��H4��20λ������Ķ�������H4K20me2���ĸ���������ļ��ʼʶ������ɸ�����ʼλ���ѡ��1��
�����Ŵ���Ϣ�ľ�ȷ���ݶ������ķ��ܺͽ���������Ҫ���ߵ��������DNA������Ҫȷ���״�DNA�������Ŵ���Ϣȷ���ݸ��Ӵ���������ʼλ�����ȷѡ�������е���Ҫ���ڡ������о��������ڶ�ϸ�������к��鵰�ױ���H2A.Z�ĺ�С��ͨ������鵰���������ת��øSUV420H1�ٽ��鵰��H4��20λ������Ķ�������H4K20me2���ĸ���������ļ��ʼʶ������ɸ�����ʼλ���ѡ��1��
����2023��8��2�գ��й���ѧԺ���������о����������������ϱ�������졢��ƽ�����飬�Լ�������ʵ��������������飬�ڡ�Molecular Cell�����߷�������ΪStructural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1���о����ģ���������ԴSUV420H1���H2A.Z��С��ĸ߷ֱ��ʵ羵�ṹ����ʾ��SUV420H1����ʶ��H2A.Z��С�岢��H4K20me2�ķ��ӻ�����ͼ1����
ͼ1��SUV420H1��H2A.Z/H2A��С�������ṹʾ��ͼ
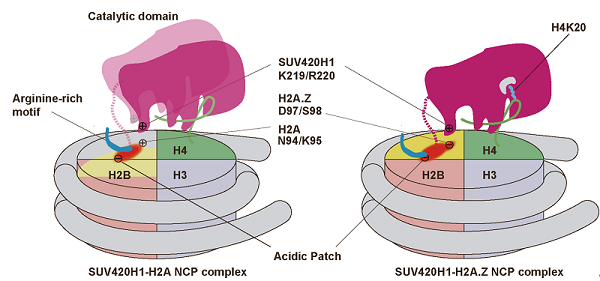
�������о�ͨ����ѧ�ϳɵķ����ڴ�λ��������������ͻ�䣬�����SUV420H1-H2A.Z��С�帴������ȶ��ԣ���������3.2 Å�Ľ�ԭ�ӷֱ��ʵ��䶳�羵�ṹ���ṹ��ʾSUV420H1���С���е�H4 N�ˡ�DNA���Լ���������Ƚ��н�ϡ����У� H4 (1-12)���С������H3-H4�������ã���H4 (13-18)��SUV420H1�ķ�϶��ϣ���ʹ��H4���췽������ת��H4 (19-24)����SUV420H1�������ġ�H4 N���γɵ���������״�ṹ��H4K20ȷ��λ��SUV420H1�����ĵ���ˮͨ����ʹ��ת�Ʒ�Ӧ��ɣ�ͼ1. B�����ƻ��ýṹ�������е���һ������ö���Ӱ��SUV420H1�ļ������ܡ��ṹ��ʾSUV420H1 KR loop��H2A.Z����л�D97/S98���ڣ���Ӧ��H2A�л�ΪN94/K95����KR loopͻ�併��SUV420H1��H2A.Z��С�壬����H2A��С��ļ������ԡ������о�����KR loopͻ�䵼��ϸ��H4K20me2ˮƽ�½��� DNA������ʼ�������Լ�ϸ������ȱ�ݡ������о��з���H2A.Z D97/S98���Ծ���SUV420H1��H2A.Z��С�������ʶ���뱾�о��Ľ��һ�¡�
�������о���ʾ��SUV420H1����ʶ��H2A.Z��С�岢������H4K20me2�Ľṹ������������SUV420H1ͨ��H2A.Z����DNA������ʼ�ķ��ӻ�����Ϊҩ���������뼲�������ṩ����Ҫ����2��H2A.Z��Ϊ�ؼ����鵰�ױ��弸������������Ⱦɫ��Ϊģ�������ѧ���̣������������������о��鵰�װ��»�Ⱦɫ�����ܸ������H2A.Z�����������ʶ��ķ��ӻ��ƣ���ʾ��H2A.Z�ں�С����װ��ȥ��װ���̵�ѡ����ʶ��ģʽ�����Ӧ������ѧ����3-8��Ȼ����H2A.Z��С����Ϊ���屻����ʶ��ķ��ӻ�����������δ�����������о��״β�����H2A.Z��С�������ʶ���������˶��鵰�ױ���ṹ���ܵ��˽⡣

����ͼ���о���ʾ�˼�ת��øSUV420H1����ʶ��H2A.Z��С�岢����H4K20me2���εĻ��ơ�SUV420H1������һ�����飬������ħ���ں��б����鵰�ĺ�С���ϵ�����ǣ����о�������H2A.Z��SUV420H1��DNA������ʼ�е����á�
�����й���ѧԺ���������о�������������졢��ƽ�о�Ա��������ʵ�����������о�ԱΪ��ͬͨѶ���ߣ����������о��������о�Ա����ʿ�����������������о�ԱΪ���ĵĹ�ͬ��һ���ߡ����о���ù�����Ȼ��ѧ����ί�������п�ѧ����ίԱ�ᡢ�й���ѧԺ��Ŀ����Ŀ���������������������ʿ�ѧƽ̨��������������ṩ����Ҫ֧�źͱ��ϡ�
�����������ӣ�
����https://www.cell.com/molecular-cell/fulltext/S1097-2765(23)00515-4
����1. Long, H., Zhang, L., Lv, M., Wen, Z., Zhang, W., Chen, X., Zhang, P., Li, T., Chang, L., Jin, C., et al. (2020). H2A.Z facilitates licensing and activation of early replication origins. Nature 577, 576-581. 10.1038/s41586-019-1877-9.
����2. Paulsen, B., Velasco, S., Kedaigle, A.J., Pigoni, M., Quadrato, G., Deo, A.J., Adiconis, X., Uzquiano, A., Sartore, R., Yang, S.M., et al. (2022). Autism genes converge on asynchronous development of shared neuron classes. Nature 602, 268-273. 10.1038/s41586-021-04358-6
����3. Huang, Y., Dai, Y., and Zhou, Z. (2020). Mechanistic and structural insights into histone H2A-H2B chaperone in chromatin regulation. Biochem. J. 477, 3367-3386. 10.1042/BCJ20190852.
����4. Dai L, Xiao X, Pan L, Shi L, Xu N, Zhang Z, Feng X, Ma L, Dou S, Wang P, Zhu B, Li W, Zhou Z. (2021) Recognition of the inherently unstable H2A nucleosome by Swc2 is a major determinant for unidirectional H2A.Z exchange. Cell Rep. 35(8):109183.
����5. Huang Y, Sun L, Pierrakeas L, Dai L, Pan L, Luk E, Zhou Z. (2020) Role of a DEF/Y motif in histone H2A-H2B recognition and nucleosome editing. Proc Natl Acad Sci USA. 117(7): 3543-3550.
����6. Wang Y, Liu S, Sun L, Xu N, Shan S, Wu F, Liang X, Huang Y, Luk E, Wu, C, Zhou Z. (2019) Structural insights into histone chaperone Chz1-mediated H2A.Z recognition and histone replacement. PLoS Biol. 17(5): e3000277.
����7. Liang X, Shan S, Pan L, Zhao J,Ranjan A, Wang F, Zhang Z, Huang Y, Feng H, Wei D, Huang L, Liu X, Zhong Q, Lou J, Li G, Wu C, Zhou Z. (2016) Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1. Nat Struct Mol Biol. 23(4):317-325.
����8. Mao Z, Pan L, Wang W, Sun J, Shan S, Dong Q, Liang X, Dai L, Ding X, Chen S, Zhang Z, Zhu B, Zhou Z. (2014) Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res. 24(4):389-399.
 ����ͨ�Ź��ں�
����ͨ�Ź��ں�
���ѣ�DNA������ʼ��|H2A.Z��С��|ѡ����ʶ��|
֪����ҵ��Ƹ